
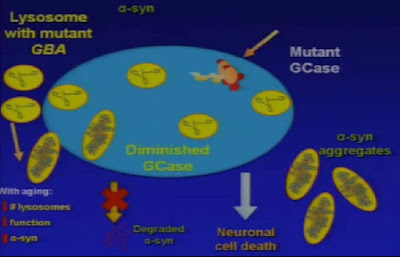
When watching the presentation on the Genetics of Parkinson's Disease by Ellen Sidransky featured yesterday. I could not but notice a few other things on top of yesterday's question and meta-question.
For one, it is abundantly clear that it is not because a metabolic circuit becomes disrupted , thanks to the lack of a certain gene, that it necessarily implies a large physiological change (also see [1]). Let us note that the robustness in random processes observed in Nature parallels some of the information preserving capacity in compressive sensing with random matrices. When people find that a specific gene is responsible for one specific disease, it really means that the culpable gene is a single point of failure. But what about those, probably more important, cases when no specific gene is a single point of failure, how do we investigate those cases in some automatized fashion (instead of just relying on chance as we have done so far) ?
Or let me put it in a different perspective that is familiar to readers of Nuit Blanche: What measurements are required for the blind deconvolution of these metabolic circuits ? I note the paucity of studies in that regard, see for instance Reverse Engineering Biochemical Networks and Compressive Sensing, It's quite simply, the stuff of Life... and Instances of Null Spaces: Can Compressive Sensing Help Study Non Steady State Metabolic Networks ?.
In the post-screening analysis, a natural extension of compressive sensing could include a better sensor and better image reconstruction for confocal imaging but I wonder if group testing and other known devices could help the other processes like it did for High Throughput Testing ?
[1] In Properties of Metabolic Networks: Structure versus Function by R. Mahadevan and B. O. Palsson on can read:
It is observed that, functionally speaking, the essentiality of reactions in a node is not correlated with node connectivity as structural analyses of other biological networks have suggested.





No comments:
Post a Comment